Table of domain abbreviations
Domains are depicted as small rectangles containing a unique domain abbreviation. Meta-domains like '7TM_chemorcpt_1', which typically cover the entire protein, are displayed as larger rectangles. The size of the rectangle does not reflect the actual domain size.
| abbreviation | display | corresponds to Pfam | corresponds to SMART | description |
|---|---|---|---|---|
| 14_3_3 | 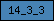 |
PF00244 | SM00101 | 14_3_3 domain |
| 3b-HSD |  |
PF01073 | 3-beta hydroxysteroid dehydrogenase/isomerase family | |
| 7TM_GPCR_Sra |  |
PF02117 | Nematode chemoreceptor, Sra | |
| 7TM_GPCR_Srab |  |
PF10292 | Serpentine type 7TM GPCR receptor class Srab | |
| 7TM_GPCR_Srb |  |
PF02175 | Serpentine beta receptor | |
| 7TM_GPCR_Srbc |  |
PF10316 | Serpentine type 7TM GPCR chemoreceptor Srbc | |
| 7TM_GPCR_Sre |  |
PF03125 | C. elegans Sre G protein-coupled chemoreceptor | |
| 7TM_GPCR_Srg |  |
PF02118 | Serpentine gamma receptor, Caenorhabditis species | |
| 7TM_GPCR_Srh |  |
PF10318 | Serpentine type 7TM GPCR receptor class Srh | |
| 7TM_GPCR_Sri |  |
PF10327 | Serpentine type 7TM GPCR receptor class Sri | |
| 7TM_GPCR_Srj |  |
PF10319 | Serpentine type 7TM GPCR receptor class Srj | |
| 7TM_GPCR_Srr |  |
PF03268 | Doamin of unknown function DUF267 | |
| 7TM_GPCR_Srsx |  |
PF10320 | Serpentine type 7TM GPCR receptor class Srsx | |
| 7TM_GPCR_Srt |  |
PF10321 | Serpentine type 7TM GPCR receptor class Srt | |
| 7TM_GPCR_Sru |  |
PF10322 | Serpentine type 7TM GPCR receptor class Sru | |
| 7TM_GPCR_Srv |  |
PF10323 | Serpentine type 7TM GPCR receptor class Srv | |
| 7TM_GPCR_Srw |  |
PF10324 | Serpentine type 7TM GPCR receptor class Srw | |
| 7TM_GPCR_Srx |  |
PF10328 | Serpentine type 7TM GPCR receptor class Srx | |
| 7TM_GPCR_Srxa |  |
PF03383 | Serpentine type 7TM GPCR receptor class Srxa | |
| 7TM_GPCR_Srz |  |
PF10325 | Serpentine type 7TM GPCR receptor class Srz | |
| 7TM_GPCR_Str |  |
PF10326 | Serpentine type 7TM GPCR receptor class Str | |
| AA_perm |  |
PF00324 | amino acid permease | |
| Aa_trans |  |
PF01490 | amino acid transporter | |
| AAA |  |
PF00004 | SM00382 | AAA ATPase |
| ABC2_m | 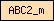 |
PF01061 | ABC-2 type transporter | |
| ABC_transporter |  |
PF00005 | ABC transporter related | |
| ABC_transporter_TM |  |
PF00664 PF06472 | ABC transporter, transmembrane region, type 1 | |
| ABHyd |  |
PF00561 | alpha/beta hydrolase fold | |
| AbHyd3 |  |
PF07859 | alpha/beta hydrolase fold | |
| ACBP |  |
PF00887 | Acyl CoA binding protein | |
| AcCoA_dh |  |
PF00441 | Acyl-CoA dehydrogenase, C-terminal domain | |
| actin |  |
PF00022 | SM00268 | Actin/actin-like |
| Acy_dh2 |  |
PF08028 | Acyl-CoA dehydrogenase, C-terminal domain | |
| Acy_dh_M |  |
PF02770 | Acyl-CoA dehydrogenase, middle domain | |
| Acy_dh_N |  |
PF02771 | Acyl-CoA dehydrogenase, N-terminal domain | |
| AcylT |  |
PF00583 | AhpC/TSA family | |
| Acyltransferase |  |
PF01757 | Acyltransferase 3 | |
| AD |  |
PF01421 | Peptidase M12B, ADAM/reprolysin | |
| ADH_N |  |
PF00107 | Zinc-binding dehydrogenase | |
| Ah_perox |  |
PF03098 | Animal haem peroxidase | |
| AhpC |  |
PF00578 | AhpC/TSA family | |
| AKR |  |
PF00248 | Aldo/keto reductase family | |
| Aldedh |  |
PF00171 | Aldehyde dehydrogenase family | |
| AMPB |  |
PF00501 | AMP-binding enzyme | |
| ANFR |  |
PF01094 | receptor family ligand binding region | |
| Ankyr |  |
PF00023 | SM00248 | Ankyrin repeat |
| APH |  |
PF01636 | Phosphotransferase enzyme family | |
| Apple |  |
PF00024 | SM00223 SM00473 | apple |
| Arm |  |
PF00514 | SM00185 | armadillo repeat |
| Arre_C |  |
PF02752 | SM01017 | Arrestin (or S-antigen), C-terminal domain |
| Arre_N |  |
PF00339 | Arrestin (or S-antigen), N-terminal domain | |
| Asp |  |
PF00026 | Aspartate protease | |
| AT |  |
PF01821 | SM00104 | Anaphylatoxin/fibulin |
| AT_12 |  |
PF00155 | Aminotransferase class I and II | |
| AT_5 |  |
PF00266 | Aminotransferase class-V | |
| AT_hook |  |
PF02178 | SM00384 | DNA binding domain with preference for A/T rich regions |
| B4_1N | 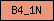 |
PF00373 | SM00295 | Band 4.1, N-terminal |
| BACK |  |
PF07707 | SM00875 | BTB And C-terminal Kelch |
| Band_7 |  |
PF01145 | SM00244 | Band 7 protein (found in stomatin) |
| Bestrophin |  |
PF01062 | Bestrophin | |
| bHLH |  |
PF00010 | SM00353 | Basic helix-loop-helix region |
| BPI2 |  |
PF02886 | SM00329 | BPI/LBP/CETP C-terminal domain |
| Branch |  |
PF02485 | Core-2/I-Branching enzyme | |
| BRCT |  |
PF00533 | SM00292 | BRCT |
| Bromo |  |
PF00439 | SM00297 | Bromodomain |
| BTB |  |
PF00651 | SM00225 | BTB/POZ-like |
| bZIP |  |
PF07716 | SM00338 | Basic-leucine zipper (bZIP) transcription factor |
| C1 | 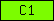 |
PF00130 | SM00109 | Protein kinase C, phorbol ester/diacylglycerol binding |
| C2 |  |
PF00168 | SM00239 | C2 calcium-dependent membrane targeting |
| C6 |  |
PF01681 PF02795 | SM01048 | Protein of unknown function C6 |
| C_AcyT |  |
PF00755 | Choline/Carnitine o-acyltransferase | |
| CA |  |
PF00028 | SM00112 | cadherin |
| Carbesterase_B |  |
PF00135 | Carboxylesterase, type B | |
| Cation_efflux |  |
PF01545 | cation efflux family | |
| CBM |  |
PF01607 | SM00494 | Peritrophin-A domain is found in chitin binding proteins |
| CBS |  |
SM00116 | Domain in cystathionine beta-synthase and other proteins | |
| CH |  |
PF00307 | SM00033 | Calponin homology domain |
| Chit |  |
SM00636 | Chitinase II | |
| CHK |  |
SM00587 | ZnF_C4 abd HLH domain containing kinases domain | |
| Chromo |  |
PF00385 | SM00298 | Chromo domain |
| CK |  |
SM00041 | Growth factor, cystine knot | |
| CL |  |
PF00059 | SM00034 | C-type lectin |
| Claudin3 |  |
PF06653 | Tight junction protein, Claudin-like | |
| CLC |  |
PF07062 | claudin homolog | |
| cNMP |  |
SM00100 | Cyclic nucleotide-monophosphate binding domain | |
| COL |  |
PF01391 | collagen | |
| ColN |  |
PF01484 | SM01088 | N-terminus of cuticular collagens |
| CRIB |  |
PF00786 | SM00285 | PAK-box/P21-Rho-binding |
| CT |  |
PF00100 | SM00241 | Endoglin/CD105 antigen |
| Ctr |  |
PF04145 | Ctr copper transporter family | |
| CtX |  |
PF02363 | C_tripleX; cysteine rich repeat | |
| CUB |  |
PF00431 PF02408 | SM00042 | CUB domain |
| CW |  |
PF08277 | SM00605 | CW domain (found in nematodes only so far ...) |
| CX |  |
PF01705 | CX domain (found in nematodes only so far ...) | |
| CY |  |
PF00031 | SM00043 | Proteinase inhibitor I25, cystatin |
| CYCL |  |
PF00134 | SM00385 | domain present in cyclins, TFIIB and Retinoblastoma |
| CysPc |  |
PF00648 | SM00230 | Calpain-like thiol protease family |
| Cyt-b5 |  |
PF00173 | Cytochrome b5 | |
| Cytochrome_P450 |  |
PF00067 | Cytochrome P450 | |
| DAO |  |
PF01266 | FAD dependent oxidoreductase | |
| DB |  |
PF01682 | DB domain (found in nematodes only so far ...) | |
| DC |  |
SM00289 | DC-domain; Cysteine-rich repeat | |
| DEAD_N |  |
SM00487 | DEAD-like helicase, N-terminal | |
| Death |  |
PF00531 | SM00005 | Death domain |
| DH_SDR |  |
PF00106 | SM00822 | Short-chain dehydrogenase/reductase SDR |
| DI |  |
PF00200 | SM00050 | disintegrin domain |
| DLH |  |
PF01738 | Dienelactone hydrolase family | |
| DM |  |
PF00751 | SM00301 | Doublesex DNA-binding motif |
| DnaJ |  |
SM00271 | DnaJ molecular chaperone homology domain | |
| DOMON |  |
PF03351 | SM00664 | DOMON domain |
| DSL |  |
PF01414 | SM00051 | Delta/Serrate/lag-2 (DSL) domain |
| DSRM |  |
PF00035 | SM00358 | Double-stranded RNA binding motif |
| DUF1096 |  |
PF06493 | Domain of unknown function DUF1096 | |
| DUF1114 |  |
PF06542 | Domain of unknown function DUF1114 | |
| DUF1248 |  |
PF06852 | Domain of unknown function DUF1248 | |
| DUF1258 |  |
PF06869 | Domain of unknown function DUF1258 | |
| DUF1261 |  |
PF06879 | Domain of unknown function DUF1261 | |
| DUF1265 |  |
PF06887 | Domain of unknown function DUF1265 | |
| DUF1280 |  |
PF06918 | Domain of unknown function DUF1280 | |
| DUF13 |  |
PF01482 | Domain of unknown function DUF13 | |
| DUF130 |  |
PF02343 | Domain of unknown function DUF130 | |
| DUF148 |  |
PF02520 | Domain of unknown function DUF148 | |
| DUF1605 |  |
PF07717 | Domain of unknown function DUF1605 | |
| DUF1647 |  |
PF07801 | Domain of unknown function 1647 | |
| DUF1679 |  |
PF07914 | Domain of unknown function DUF1679 | |
| DUF19 |  |
PF01579 PF03236 | Domain of unknown function DUF19 | |
| DUF227 |  |
PF02958 | Domain of unknown function DUF227 | |
| DUF229 |  |
PF02995 | Domain of unknown function DUF229 | |
| DUF23 |  |
PF01697 | Domain of unknown function DUF23 | |
| DUF236 |  |
PF03057 | Domain of unknown function DUF236 | |
| DUF2650 |  |
PF10853 | Domain of unknown function DUF2650 | |
| DUF268 |  |
PF03269 | Domain of unknown function DUF268 | |
| DUF271 |  |
PF03407 | Domain of unknown function DUF271 | |
| DUF272 |  |
PF03312 | Domain of unknown function DUF272 | |
| DUF273 |  |
PF03314 | Domain of unknown function DUF273 | |
| DUF274 |  |
PF03409 | Domain of unknown function DUF274 | |
| DUF281 |  |
PF03436 | Domain of unknown function DUF281 | |
| DUF282 |  |
PF03380 | Domain of unknown function DUF282 | |
| DUF288 |  |
PF03385 | Domain of unknown function DUF288 | |
| DUF316 |  |
PF03761 | Domain of unknown function DUF316 | |
| DUF3557 |  |
PF12078 | Domain of unknown function DUF3557 | |
| DUF38 |  |
PF01827 | Domain of unknown function DUF38 | |
| DUF595 |  |
PF04590 | Domain of unknown function DUF595 | |
| DUF621 |  |
PF04789 | Domain of unknown function DUF621, putative G-protein coupled receptor | |
| DUF644 |  |
PF04870 | Domain of unknown function DUF644 | |
| DUF672 |  |
PF05050 | Domain of unknown function DUF672 | |
| DUF684 |  |
PF05075 | Domain of unknown function DUF684 | |
| DUF780 |  |
PF05611 | Domain of unknown function DUF780 | |
| DUF870 |  |
PF05912 | Domain of unknown function DUF870 | |
| DUF976 |  |
PF06162 | Domain of unknown function DUF976 | |
| DUF_CC |  |
PF04942 | Domian of unknown function, DUF-CC | |
| E1E2ATP |  |
PF00122 | proton ATPase | |
| EamA |  |
PF00892 | EamA-like transporter family | |
| EB |  |
PF01683 | EB-domain (found in nematodes only so far ...) | |
| ECH |  |
PF00378 | Enoyl-CoA hydratase/isomerase family | |
| EF-Tu |  |
PF00009 | GTP-binding elongation factor family, EF-Tu/EF-1A subfamily | |
| EF-Tu2 |  |
PF03144 | Elongation factor Tu domain 2 | |
| EF_hand |  |
PF00036 | SM00054 | Calcium-binding EF-hand |
| EGF |  |
PF00008 PF07974 PF07645 | SM00001 SM00179 SM00181 | EGF module |
| Epi |  |
PF01370 | NAD dependent epimerase/dehydratase family | |
| ET |  |
PF01684 | Domain of unknown function ET | |
| Ets |  |
PF00178 | SM00413 | Ets domain |
| EXOIII |  |
SM00479 | exonuclease domain in DNA-polymerase alpha and epsilon chain, ribonuclease T and other exonucleases | |
| F5_8C |  |
PF00754 | SM00231 | Coagulation factor 5/8 type, C-terminal |
| FADB2 |  |
PF00890 | FAD binding domain | |
| FARP |  |
PF01581 | FMRFamide related peptide family | |
| Fba_2 |  |
PF07735 | F-box associated type 2 | |
| Fbox |  |
PF00646 | SM00256 | Fbox |
| FeoB_N |  |
PF02421 | Ferrous iron transport protein B | |
| FERM_C |  |
PF09380 | FERM C-terminal PH-like domain | |
| FG |  |
PF00147 | SM00186 | Fibrinogen, alpha/beta/gamma chain, C-terminal globular |
| FHA |  |
PF00498 | SM00240 | Forkhead-associated |
| FN3 |  |
PF00041 | SM00060 | Fibronectin, type III-like fold |
| FolN |  |
PF09120 PF09289 | SM00274 | Follistatin-like, N-terminal |
| Fork_Hd |  |
PF00250 | SM00339 | Fork head transcription factor |
| Fringe |  |
PF02434 | Fringe-like | |
| FU |  |
SM00261 | furin repeat | |
| Fukutin |  |
PF06828 | Fukutin-related | |
| FZ |  |
PF01392 | SM00063 | Frizzled cysteine-rich domain |
| G_alpha |  |
PF00503 | SM00275 | Guanine nucleotide binding protein (G-protein), alpha subunit |
| G_patch |  |
PF01585 | SM00443 | glycine rich nucleic binding domain |
| Gal_T |  |
PF01762 | Galactosyltransferase | |
| Gcyc |  |
PF00211 | SM00044 | guanylate cyclase |
| GL |  |
PF00337 | SM00276 SM00908 | Galectin, galactose-binding lectin |
| Globin |  |
PF00042 | Globin-like | |
| Glycoside_hydrolase |  |
PF00933 | Glycoside hydrolase, catalytic core | |
| GlyT |  |
PF01531 | Glycosyl transferase family 11 | |
| GlyT28_C |  |
PF04101 | Glycosyltransferase family 28 C-terminal domain | |
| GlyTr2 |  |
PF00535 | Glycosyl transferase family 2 | |
| GPCR_Rhodopsin |  |
PF00001 | GPCR, rhodopsin-like superfamily | |
| GPCR_secretin |  |
PF00002 | GPCR, family 2, secretin-like | |
| GPS |  |
PF01825 | SM00303 | GPS domain |
| GR |  |
PF00462 | Glutaredoxin | |
| Gra6 |  |
PF05084 | Gra6 domain | |
| Gran |  |
PF00396 | SM00277 | Granulin |
| Grd |  |
PF04155 | ground domain | |
| GST_N |  |
PF02798 | Glutathione S-transferase, N-terminal domain | |
| GTPase |  |
PF00025 PF00071 PF08477 | SM00010 SM00173 SM00174 SM00175 SM00176 SM00177 SM00178 | small GTPases (all subfamilies, Ras, Ran, Rac, etc...) |
| H2A |  |
SM00414 | histone H2A | |
| H2B |  |
SM00427 | histone H2B | |
| H3 |  |
SM00428 | histone H3 | |
| H4 |  |
SM00417 | histone H4 | |
| HA2 |  |
PF04408 | SM00847 | Helicase associated domain (HA2) |
| HEAT |  |
PF02985 | HEAT | |
| Helic_C |  |
PF00271 | SM00490 | DNA/RNA helicase, C-terminal |
| HintC |  |
SM00305 | Hint (Hedgehog/Intein) domain C-terminal region | |
| HintN |  |
PF01079 | SM00306 | Hint (Hedgehog/Intein) domain N-terminal region |
| HisPhos2 |  |
PF00328 | Histidine phosphatase superfamily (branch 2) | |
| Histone |  |
PF00125 PF00538 | SM00526 | Histone fold |
| HMG |  |
PF00505 | SM00398 | High mobility group box, HMG1/HMG2, subgroup |
| HOM |  |
PF00046 | SM00389 | homeobox |
| HSP20 |  |
PF00011 | HSP20-like chaperone | |
| Hsp70 |  |
PF00012 | Heat shock protein 70 | |
| HX |  |
PF00045 | SM00120 | hemopexin |
| Hydrol |  |
PF00702 | haloacid dehalogenase-like hydrolase | |
| I29 |  |
SM00848 | Cathepsin propeptide inhibitor domain (I29) | |
| IBR |  |
PF01485 | SM00647 | In Between Ring fingers |
| IFP |  |
PF00038 | Intermediate filament protein | |
| IG |  |
PF07654 PF07679 PF07686 PF00047 PF08205 | SM00406 SM00407 SM00408 SM00409 SM00410 | immunoglobulin domain |
| Innexin |  |
PF00876 | Innexin | |
| Insulin |  |
PF03488 | SM00078 | Nematode insulin-related peptide, beta type |
| Ion_glu_rcpt |  |
PF00060 | Ionotropic glutamate receptor | |
| Ion_tr |  |
PF00520 | four TM helices additionally present in many channels | |
| IQ |  |
PF00612 | SM00015 | Short calmodulin-binding motif containing conserved Ile and Gln residues |
| JmjC |  |
PF02373 PF08007 | SM00558 | A domain family that is part of the cupin metalloenzyme superfamily |
| K_chan_2pore |  |
PF07885 | Potassium channel, two pore-domain | |
| K_chan_volt |  |
PF02214 | Potassium channel, voltage dependent, Kv, tetramerisation | |
| Kelch |  |
PF01344 PF07646 | SM00612 | Kelch repeat type 1 |
| KH |  |
PF00013 | SM00322 | K Homology |
| KIN |  |
PF00069 PF07714 | SM00219 SM00220 SM00221 | protein kinase |
| Kin_Ct |  |
SM00133 | AGC-kinase, C-terminal | |
| Kinesin_motor |  |
PF00225 | SM00129 | Kinesin, motor region |
| KR |  |
PF00051 | SM00130 | kringle |
| KU |  |
PF00014 | SM00131 | Kunitz domain/Proteinase inhibitor I2 |
| KZ |  |
PF00050 PF07648 | SM00280 | Proteinase inhibitor I1, Kazal |
| LA |  |
PF00057 | SM00192 | Low density lipoprotein-receptor, class A domain |
| Lactam_B |  |
PF00753 | SM00849 | Metallo-beta-lactamase superfamily |
| LamB |  |
PF00052 | SM00281 | Laminin B domain |
| LE |  |
PF00053 | SM00180 | laminin EGF-like domain |
| LG |  |
PF00054 PF02210 | SM00210 SM00282 | laminin G domain |
| LGC-GluB |  |
PF10613 | SM00918 | Ligated ion channel L-glutamate- and glycine-binding site |
| LIM |  |
PF00412 | SM00132 | LIM domain |
| Lin-8 |  |
PF03353 | Ras-mediated vulval-induction antagonist | |
| Lip_2 |  |
PF01674 | Lipase (class 2) | |
| Lip_3 |  |
PF01764 | Lipase (class 3) | |
| Lip_G |  |
PF00657 | GDSL-like Lipase/Acylhydrolase | |
| LisH |  |
SM00667 | Lissencephaly type-1-like homology motif | |
| LITAF |  |
SM00714 | Possible membrane-associated motif in LPS-induced tumor necrosis factor alpha factor | |
| LN |  |
PF00055 | SM00136 | laminin, N-terminal domain |
| LRR |  |
PF00560 | SM00369 SM00370 SM00368 | leucine-rich repeat |
| LRRc |  |
PF01463 | SM00082 | leucine-rich repeat, C-terminal |
| LRRn |  |
PF01462 | SM00013 | leucine-rich repeat, N-terminal |
| LTD |  |
PF00932 | Lamin Tail Domain | |
| LY |  |
PF00058 | SM00135 | Low-density lipoprotein receptor, YWTD repeat |
| LZ |  |
PF04916 | laminin A domain | |
| MAM |  |
PF00629 | SM00137 | MAM domain |
| MATH |  |
PF00917 | SM00061 | MATH domain |
| MBOAT |  |
PF03062 | membrane-bound o-acyltransferase | |
| MD |  |
SM00604 | MD domain (found in nematodes only so far ...) | |
| MFP2b |  |
PF12150 | Cytosolic motility protein | |
| MFS_1 |  |
PF07690 | Major facilitator superfamily MFS-1 | |
| Mitoch_carrier |  |
PF00153 | Mitochondrial substrate carrier | |
| MMR |  |
PF01926 | 50S ribosome-binding GTPase | |
| MP |  |
SM00235 | metalloprotease (covers all, should be replaced in display by the appropriate subfamily, e.g. PepM12) | |
| MreB |  |
PF06723 | MreB/Mbl protein | |
| MSP |  |
PF00635 | Major sperm protein | |
| MSP4 |  |
PF07993 | Male sterility protein | |
| MT_11 |  |
PF08241 | Methyltransferase domain | |
| MT_12 |  |
PF08242 | Methyltransferase domain | |
| MTS |  |
PF05175 | Methyltransferase small domain | |
| MYND |  |
PF01753 | MYND finger | |
| Myosin_head |  |
PF00063 | SM00242 | Myosin head, motor region |
| Myosin_tail |  |
PF01576 | Myosin tail | |
| N1 |  |
PF06119 | SM00539 | nidogen, N-terminal domain |
| Na_Ca_ex |  |
PF01699 | sodium/calcium exchanger protein | |
| Na_chan_AS |  |
PF00858 | Na+ channel, amiloride-sensitive | |
| Na_diCO_symport |  |
PF00375 | Sodium:dicarboxylate symporter | |
| Na_H_ex |  |
PF00999 | sodium/hydrogen exchanger family | |
| NC10 |  |
PF06482 | endostatin | |
| Neuro_channel_lgbd |  |
PF02931 | Neurotransmitter-gated ion-channel | |
| Neuro_channel_tm |  |
PF02932 | Neurotransmitter-gated ion-channel transmembrane region | |
| NhrLig |  |
PF00104 | SM00430 | Nuclear hormone receptor, ligand-binding |
| NRF |  |
SM00703 | N-terminal domain in C. elegans NRF-6 (Nose Resistant to Fluoxetine-4) and NDG-4 (resistant to nordihydroguaiaretic acid-4) | |
| Nuclease |  |
PF00929 | Polynucleotidyl transferase, Ribonuclease H fold | |
| NucST |  |
PF04142 | Nucleotide-sugar transporter | |
| NUDIX |  |
PF00293 | NUDIX domain | |
| P-Diest |  |
PF01663 | Type I phosphodiesterase / nucleotide pyrophosphatase | |
| P_ATP_C |  |
PF00689 | Cation transporting ATPase, C-terminus | |
| P_ATP_N |  |
SM00831 | Cation transporter/ATPase, N-terminus | |
| P_Propr |  |
PF01483 | Proprotein convertase P-domain | |
| PALP |  |
PF00291 | Pyridoxal-phosphate dependent enzyme | |
| Patatin |  |
PF01734 | Patatin-like phospholipase | |
| Patched |  |
PF02460 | Patched | |
| PAW |  |
PF04721 | SM00613 | Protein of unknown function PAW |
| PAX |  |
PF00292 | SM00351 | Paired box protein, N-terminal |
| PAZ |  |
PF02170 | SM00949 | Argonaute and Dicer protein, PAZ |
| PBPb |  |
PF00497 | SM00062 | Bacterial periplasmic substrate-binding proteins |
| PBPe |  |
SM00079 | Eukaryotic homologues of bacterial periplasmic substrate binding proteins | |
| PDZ |  |
PF00595 | SM00228 | PDZ domain |
| PepC1 |  |
SM00645 | Peptidase family C1 | |
| PepM1 |  |
PF01433 | Peptidase family M1 | |
| PepM10 |  |
PF00413 | matrixin and adamalysin; Peptidase M10A and M12B | |
| PepM12 |  |
PF01400 | astacin, peptidase M12A | |
| PepM13 |  |
PF01431 | neprilysin | |
| PepM13_N |  |
PF05649 | Peptidase family M13 | |
| PepM14 |  |
PF00246 | SM00631 | Peptidase M14, carboxypeptidase A |
| PepM16 |  |
PF00675 | Insulinase (Peptidase family M16) | |
| PepM16_C | 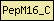 |
PF05193 | Peptidase M16 inactive domain | |
| PepS1 |  |
PF00089 | SM00020 | Peptidase S1 and S6, chymotrypsin/Hap; Peptidase, trypsin-like serine and cysteine |
| PepS10 |  |
PF00450 | Peptidase family S10 | |
| PepS28 |  |
PF05577 | Serine carboxypeptidase S28 | |
| PepS8 |  |
PF00082 | Peptidase S8 and S53, subtilisin, kexin, sedolisin | |
| PepS9 |  |
PF00326 | Prolyl oligopeptidase family | |
| Pfam |  |
all other Pfam domains | ||
| PGAM |  |
PF00300 | SM00855 | Phosphoglycerate mutase family |
| PH |  |
PF00169 PF00568 PF00640 | SM00233 SM00461 SM00462 | Pleckstrin-homology domain |
| Phox |  |
PF00787 | SM00312 | Phox-like |
| Phtase |  |
PF00149 | SM00156 | phosphoprotein phosphatase |
| PI3K |  |
PF00794 PF00454 | SM00144 SM00146 | Phosphatidylinositol 3-kinase |
| PINT |  |
PF01399 | SM00088 | motif in proteasome subunits, Int-6, Nip-1 and TRIP-15 |
| Piwi |  |
PF02171 | SM00950 | Stem cell self-renewal protein Piwi |
| PKD |  |
PF08016 | Polycystin cation channel | |
| PlsC | 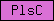 |
SM00563 | Phosphate acyltransferases | |
| Pmp3 |  |
PF01679 | proteolipid membrane potential modulator | |
| Polys2 | 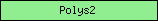 |
PF02719 | Polysaccharide biosynthesis protein | |
| POU |  |
PF00157 | SM00352 | POU domain |
| PP2Cc |  |
PF00481 PF07228 | SM00331 SM00332 | 1) Sigma factor PP2C-like phosphatases, 2) Serine/threonine phosphatases, family 2C, catalytic domain |
| Prefoldin |  |
PF01920 | Prefoldin | |
| PrmA |  |
PF06325 | Ribosomal protein L11 methyltransferase (PrmA) | |
| Pro_iso |  |
PF00160 | Cyclophilin type peptidyl-prolyl cis-trans isomerase/CLD | |
| Protea |  |
PF00227 | Proteasome subunit | |
| PTP |  |
PF00102 PF00782 | SM00012 SM00194 SM00195 SM00404 | protein-tyrosine phosphatase |
| Ptre |  |
PF00088 | SM00018 | P-type trefoil domain |
| PUF |  |
PF00806 | SM00025 | Pumilio-like repeats |
| Pyr_deC |  |
PF00282 | Pyridoxal-dependent decarboxylase conserved domain | |
| PyrR2 |  |
PF07992 | Pyridine nucleotide-disulphide oxidoreductase | |
| Ras_as |  |
PF00788 | SM00314 | ras-association domain |
| RasGAP |  |
PF00616 | SM00323 | ras GTPase activating protein |
| RcpL |  |
PF01030 | EGF receptor, L domain | |
| Redox |  |
PF08534 | Redoxin | |
| RGS |  |
PF00615 | SM00315 | Regulator of G protein signalling |
| RHOD |  |
PF00581 | SM00450 | Rhodanese Homology Domain |
| RhoGAP |  |
PF00620 | SM00324 | RhoGAP domain |
| RhoGEF |  |
PF00621 | SM00325 | RhoGEF domain |
| Rhomb |  |
PF01694 | Rhomboid protease | |
| RICIN |  |
SM00458 | Ricin-type beta-trefoil | |
| RmSB |  |
PF04321 | RmlD substrate binding domain | |
| RRM |  |
PF00076 | SM00360 SM00361 SM00362 | RNA recognition motif, RNP-1 |
| S1 |  |
PF00575 | SM00316 | Ribosomal protein S1-like RNA-binding domain |
| SAM |  |
PF00536 PF07647 | SM00454 | Sterile alpha motif homology |
| SANT |  |
PF00249 | SM00717 | SANT SWI3, ADA2, N-CoR and TFIIIB'' DNA-binding domains |
| SapB |  |
SM00741 | Saposin B | |
| SCPlike |  |
PF00188 | SCP-like extracellular | |
| SEA |  |
PF01390 | SM00200 | SEA domain |
| SEC14 |  |
PF00650 | SM00516 | Domain in homologues of a S. cerevisiae phosphatidylinositol transfer protein (Sec14p) |
| SERPIN |  |
PF00079 | SM00093 | SERine Proteinase Inhibitors |
| SET |  |
PF00856 | SM00317 | SET domain |
| SH2 |  |
PF00017 | SM00252 | SH2 domain |
| SH3 |  |
PF00018 | SM00326 | SH3 domain |
| ShK |  |
PF01549 | SM00254 | Metridin-like ShK toxin |
| Skp1 |  |
SM00512 | Found in Skp1 protein family | |
| Sm |  |
PF01423 | SM00651 | snRNP Sm proteins |
| SM |  |
PF01437 | SM00423 | semaphorin/plexin domain |
| SMART | 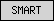 |
all other SMART domains | ||
| SNF |  |
PF00209 | sodium neurotransmitter symporter | |
| SNF2_N |  |
PF00176 | SNF2-related | |
| Snf7 |  |
PF03357 | Snf7 | |
| SP |  |
signal peptide | ||
| Spec |  |
PF00435 | SM00150 | spectrin repeat |
| SPK |  |
PF04435 | SM00583 | SPK is a domain of unknown function found in SET and PHD domain containing proteins |
| SPRY |  |
PF00622 | SM00449 | Domain in SPla and the RYanodine Receptor. |
| SR |  |
PF00530 | SM00202 | Speract/scavenger receptor |
| Ster-S |  |
PF12349 | Sterol-sensing domain of SREBP cleavage-activation | |
| Sugar_tr |  |
PF00083 | sugar transporter | |
| Sushi |  |
PF00084 | SM00032 | Sushi/SCR/CCP domain |
| SynN |  |
SM00503 | Syntaxin N-terminal domain | |
| T1 |  |
PF00090 | SM00209 | Thrombospondin, type I |
| T_Box |  |
PF00907 | SM00425 | Transcription factor, T-box |
| TBC |  |
PF00566 | SM00164 | Domain in Tre-2, BUB2p, and Cdc16p. Probable Rab-GAPs. |
| TCP1 |  |
PF00118 | HSP60 | |
| Tetraspanin |  |
PF00335 | tetraspanin | |
| TGFb |  |
PF00019 | SM00204 | TGF beta domain |
| TIL |  |
PF01826 | prot. Inhibitor domain | |
| TM |  |
transmembrane domain | ||
| TPR |  |
PF00515 PF07719 | SM00028 | Tetratricopeptide region |
| TPT |  |
PF03151 | Triose-phosphate Transporter family | |
| Transthyr_rel |  |
PF01060 | Transthyretin-like | |
| Tro_My |  |
PF00261 | Tropomyosin | |
| tSNARE |  |
SM00397 | helical region found in SNAREs | |
| Tubulin_Cterm |  |
PF03953 | SM00865 | Tubulin/FtsZ, C-terminal |
| Tubulin_GTPase |  |
PF00091 | SM00864 | Tubulin/FtsZ, GTPase |
| TY |  |
PF00086 | SM00211 | Thyroglobulin type-1 domain |
| UAA |  |
PF08449 | UAA transporter family | |
| Ub_MT |  |
PF01209 | ubiE/COQ5 methyltransferase family | |
| UBA |  |
PF00627 | SM00165 | Ubiquitin associated domain |
| UBCc |  |
SM00212 | Ubiquitin-conjugating enzyme E2, catalytic domain | |
| Ubiq |  |
PF00240 | SM00213 | Ubiquitin |
| UBQ_conjugat_E2 |  |
PF00179 | Ubiquitin-conjugating enzyme/RWD-like | |
| UCH |  |
PF00443 | Ubiquitin carboxyl-terminal hydrolase | |
| UDP_gluc_trans |  |
PF00201 | UDP-glucuronosyl/UDP-glucosyltransferase | |
| UL |  |
PF02140 | D-galactoside lectin | |
| UNC-93 | 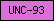 |
PF05978 | Ion channel regulatory protein UNC-93 | |
| V5_TPX |  |
SM00198 | Allergen V5/Tpx-1 related | |
| VA |  |
PF00092 | SM00327 | von Willebrand factor, type A domain |
| VC | 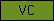 |
PF00093 | SM00214 SM00215 | von Willebrand factor, type C domain |
| VD |  |
PF00094 | SM00216 | von Willebrand factor, type D domain |
| Vincul |  |
PF01044 | Vinculin/alpha-catenin | |
| VOMI |  |
PF03762 | Vitelline membrane outer layer protein I (VOMI) | |
| WA |  |
PF00095 | SM00217 | Whey acidic protein, 4-disulphide core |
| WD40 |  |
PF00400 | SM00320 | WD40/YVTN repeat |
| Wilms |  |
PF02165 | Wilm's tumour protein | |
| WSN | 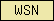 |
PF02206 | SM00453 | WSN domain (found in nematodes only so far ...) |
| WW |  |
PF00397 | SM00456 | WW/Rsp5/WWP |
| Zip |  |
PF02535 | zinc transporter | |
| Znf |  |
PF01428 PF01529 PF02135 PF01363 PF00643 PF00569 PF00642 PF00320 PF00105 PF00641 PF00098 PF00628 PF00096 | SM00064 SM00154 SM00184 SM00249 SM00251 SM00291 SM00336 SM00343 SM0352 SM00355 SM00356 SM00399 SM00401 SM00451 SM00547 SM00551 SM00744 | zinc finger |
| ZU5 | 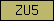 |
PF00791 | SM00218 | ZU5 domain |